Episode Ascertainment
Introduction
In EHR databases, prescriptions are usually provided as a single row for each record for every individual.
These need to be merged to form a treatment episode (Figure 1, Panel C).
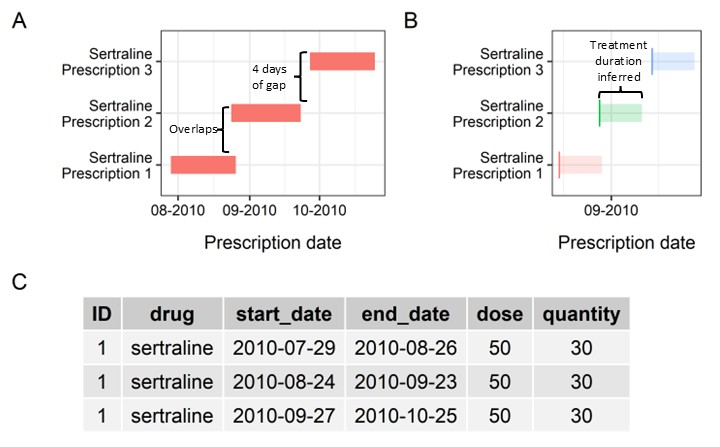
Figure 1: Sertraline sample prescriptions
One common approach is to construct treatment episodes using continuous prescription records – two prescriptions can be assigned to the same treatment episode if the periods of coverage overlap (Figure 1, Panel A).
In some EHR databases, the duration of exposure might be unavailable, and assumptions are needed to infer treatment episodes.
A simple example is shown here, assuming all prescriptions last for 28 days (Figure 1, Panel B).
T-Rx provides options to merge prescriptions into treatment episodes as longitudinal periods of exposure by:
| Function | Condition |
|---|---|
rx_merge |
when coverage (start and end date) of each prescription is available |
rx_infer |
when coverage of each prescription is not available |
Sample dataframe
In this tutorial, we utilized the data object attached with T-Rx
object, rx_demo_1 for illustration.
It is a hypothetical prescription dataframe consists of 35 rows and 8 columns (for 4 patients), with details described below.
Column name descriptions
| Column | Details |
|---|---|
ID |
Numeric identifier for individual patients |
drug |
Name of the prescribed drug |
class |
Drug class or category (e.g., SSRI) |
start_date |
Start date of the prescription |
end_date |
End date of the prescription |
dose |
Dosage of the prescribed drug |
dose_unit |
Dosage unit of the prescribed drug |
quantity |
Quantity of the drug prescribed in the prescription |
rx_merge() - Merge Prescriptions into Exposure
Periods
Description
Consider the sertraline prescriptions on the right (Panel A/B).
The first two prescriptions are in overlap, while the start date of third one has a 4-day gap with the end date of the previous prescription.
rx_merge() aggregates them into the same treatment
episode, if the coverages of prescriptions overlap (Panel
C/D).
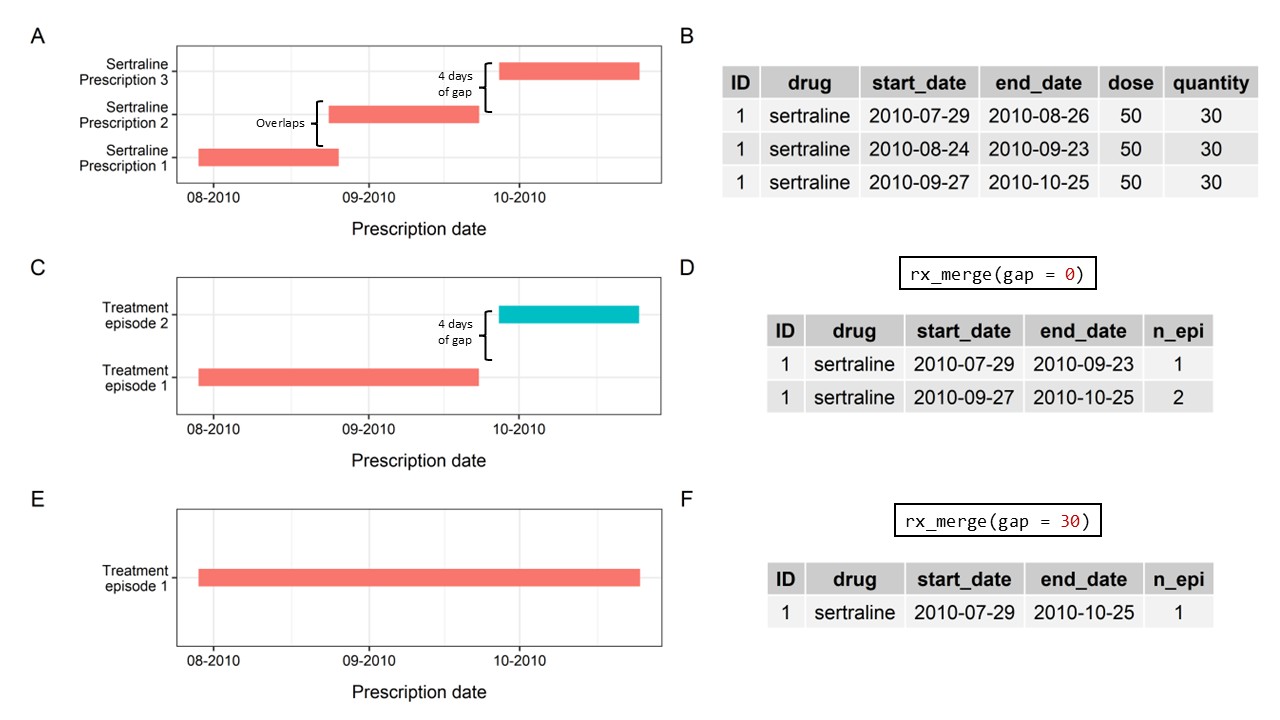
Figure 2: rx_merge function
rx_merge() also allows a window of gap between
prescriptions to account for real-world treatment complexity, such as
nudges in follow-up periods due to medication stockpiling, and avoid
misclassification as treatment episode discontinuation.
This window can be tweaked by changing the gap argument
in rx_merge() function.
With a 7-day gap, prescriptions that were less than 7 days apart would be merged together into the same treatment episode (Panel E/F).
Example & Expected Input
rx_merge() allows users to merge prescriptions based on
two major options, specified by
merge_option.
| Option | Condition |
|---|---|
date |
By differences between prescription dates (when the start and end dates of prescriptions are available) |
duration |
By dates and duration of prescriptions (when the dates and durations of prescriptions are available) |
The below demonstration of rx_merge() focuses on using
the date option.
This is the more commonly used option in most EHR databases, since the dates of prescriptions are more readily available.
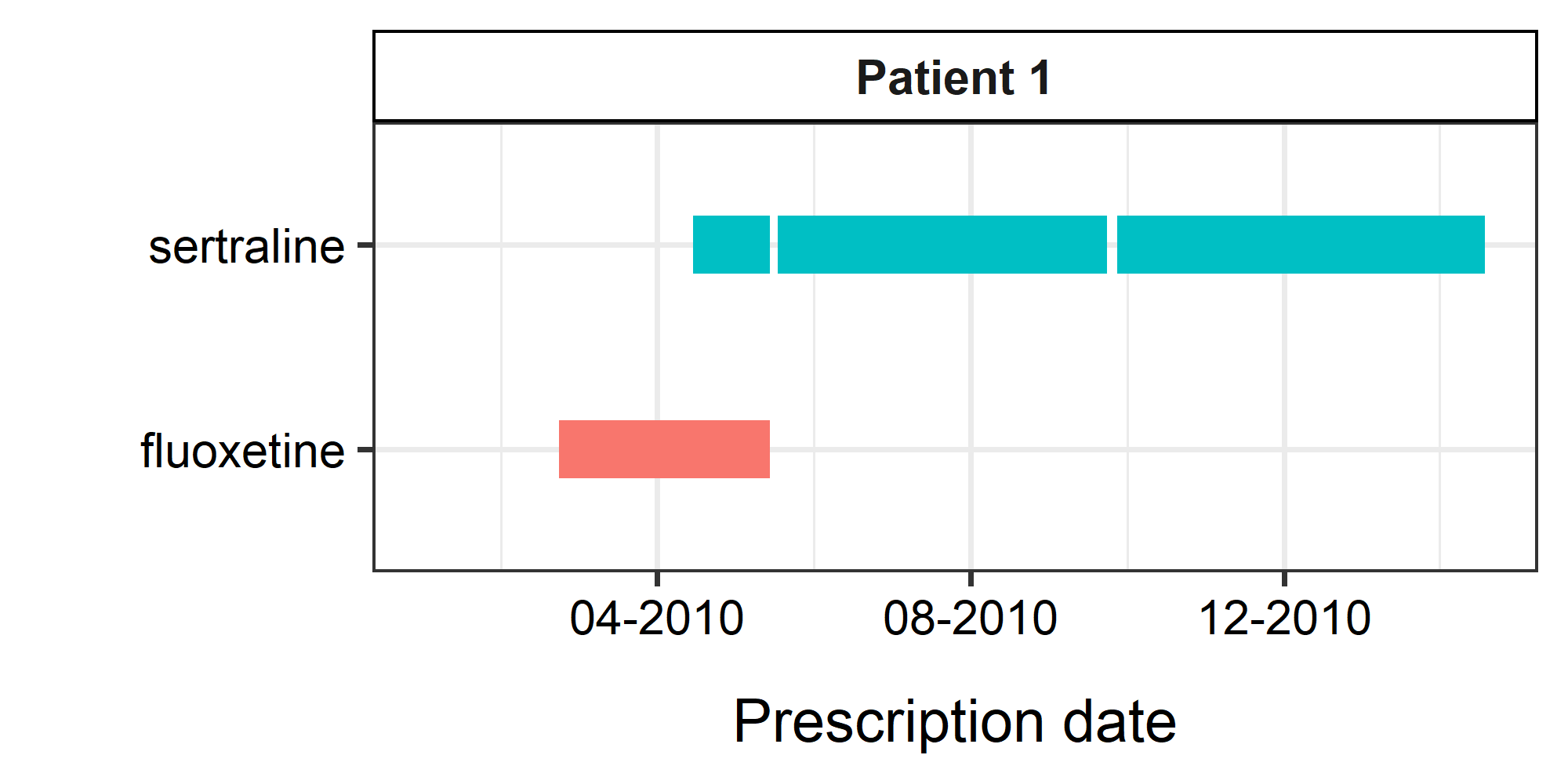
Visualizing the prescriptions
In the rx_demo_1 data object, the start date and end
dates were available as start_date and
end_date columns in the dataframe.
These prescriptions were visualized as shown on the right.
We have limited the prescriptions to those prescribed after 2010-01-01, for the ease of illustration.
Description of the data object
(rx_demo_1) can also be found
here.
Instructions
Firstly, try an initial run with the data object.
rx_date = rx_merge(rx_df = rx_demo_1,
merge_option = "date", gap = 30,
id_col = "ID", rx_date_col = "start_date", rx_end_col = "end_date",
drug_group = c("drug"))The above code specified a 30-day gap between prescriptions (from
gap option).
This means that the prescriptions would be treated as a continuous period of exposure if the following prescription is made within 30 days from the end date of previous prescription.
Explanation on arguments
| Argument | Details |
|---|---|
rx_df |
The prescription dataframe
(rx_demo_1) |
merge_option |
Merge the prescriptions based on prescription dates |
id_col |
Column name that contains patient ID
(ID) |
rx_date_col |
Column name that contains the (start) dates of
prescriptions (start_date) |
rx_end_col |
Column name that contains the end dates of
prescriptions (end_date) |
drug_group |
A vector of column names in which the prescriptions
were merged based on. We use drug here as we want to
merge prescriptions belonging to the same drug name
(drug) |
Output Dataframe
Running rx_merge() returns a dataframe of treatment
episodes of 4 patients (10 rows):
| ID | drug | start_date | end_date | n_epi |
|---|---|---|---|---|
| 1 | fluoxetine | 2010-02-22 | 2010-05-15 | 1 |
| 1 | sertraline | 2010-04-15 | 2011-02-17 | 1 |
| 2 | amitriptyline | 2007-01-30 | 2007-04-30 | 1 |
| 2 | amitriptyline | 2010-02-14 | 2010-05-15 | 2 |
| 3 | paroxetine | 2010-01-01 | 2010-05-17 | 1 |
| 4 | amitriptyline | 2010-09-30 | 2010-10-24 | 1 |
| 4 | citalopram | 2009-12-24 | 2010-03-27 | 1 |
| 4 | fluoxetine | 2010-02-25 | 2010-05-20 | 1 |
| 4 | perphenazine | 2010-09-30 | 2010-10-24 | 1 |
| 4 | venlafaxine | 2010-05-22 | 2010-09-25 | 1 |
Details of dataframe
| Column | Details |
|---|---|
ID |
Patient ID (same as input dataframe) |
drug |
Medication (same as input dataframe) |
start_date |
Start date of the treatment episode (1st prescription) |
end_date |
End date of the treatment episode (final prescription) |
n_epi |
Number of episode within the group
(drug) |
As a comparison, we have put in the dataframes for patient 1 before
and after running rx_merge():
Before rx_merge()
| ID | drug | class | start_date | end_date | dose | dose_unit | quantity |
|---|---|---|---|---|---|---|---|
| 1 | fluoxetine | SSRI | 2010-02-22 | 2010-03-22 | 20 | mg | 28 |
| 1 | fluoxetine | SSRI | 2010-03-22 | 2010-04-19 | 20 | mg | 50 |
| 1 | sertraline | SSRI | 2010-04-15 | 2010-05-15 | 50 | mg | 14 |
| 1 | fluoxetine | SSRI | 2010-04-15 | 2010-05-15 | 20 | mg | 14 |
| 1 | sertraline | SSRI | 2010-05-18 | 2010-06-15 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-06-10 | 2010-07-04 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-07-02 | 2010-08-01 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-07-29 | 2010-08-26 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-08-24 | 2010-09-23 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-09-27 | 2010-10-25 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-10-25 | 2011-01-23 | 50 | mg | 90 |
| 1 | sertraline | SSRI | 2011-01-20 | 2011-02-17 | 50 | mg | 30 |
After rx_merge()
| ID | drug | start_date | end_date | n_epi |
|---|---|---|---|---|
| 1 | fluoxetine | 2010-02-22 | 2010-05-15 | 1 |
| 1 | sertraline | 2010-04-15 | 2011-02-17 | 1 |
All sertraline / fluoxetine prescriptions were merged together after
running rx_merge().
Customizing parameters
As in Figure 1, Panel C/D, a customizable gap
between prescriptions can be specified by users to account for real-life
treatment complexities, using the gap
option in rx_merge().
If gap is specified as 0,
prescriptions with any gaps (as shown on the right) in between would be
treated as separate treatment episodes.
Run rx_merge() again, but change
gap as 0.
rx_date = rx_merge(rx_df = rx_demo_1,
merge_option = "date", gap = 0,
id_col = "ID", rx_date_col = "start_date", rx_end_col = "end_date",
drug_group = c("drug", "dose"))Note: If users want to construct treatment episodes
based on more than one criteria (i.e.: not just same drug, but also same
dose etc.), these groups can be specified using the
drug_group option in
rx_merge(). The resultant dataframe would also show these
groups as additional columns (dose in this case).
The resultant dataframe would be shown as below (for patient 1).
| ID | drug | start_date | end_date | n_epi |
|---|---|---|---|---|
| 1 | fluoxetine | 2010-02-22 | 2010-05-15 | 1 |
| 1 | sertraline | 2010-04-15 | 2010-05-15 | 1 |
| 1 | sertraline | 2010-05-18 | 2010-09-23 | 2 |
| 1 | sertraline | 2010-09-27 | 2011-02-17 | 3 |
rx_infer() - Making Inference on periods of
exposure
Description
The end dates of prescriptions are not always available in EHR databases, such as UK Biobank.
As a result, the coverage of prescriptions cannot be ascertained and
rx_merge() therefore cannot be applied.
In rx_merge(), T-Rx infers treatment duration and
constructs treatment episodes based on blocks of “repeated
prescriptions”, defined from consecutive prescriptions that were:
- of the same drug;
- of the same dosage (optional);
- of the same frequency or quantity (optional); and
- close enough in prescription dates (specified by
rx_window_daysargument).
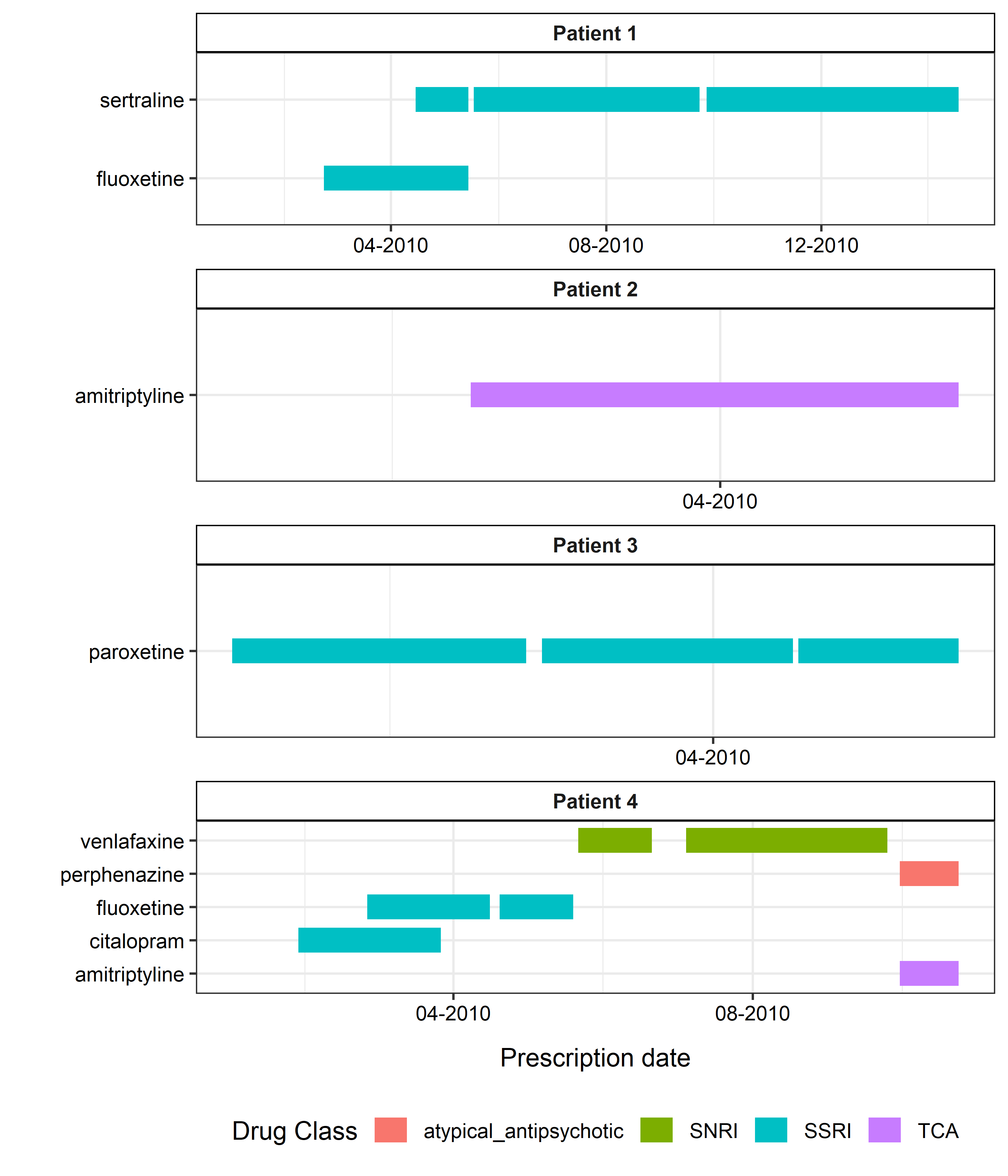
Repeated Prescriptions
A sample figure the fluoxetine prescriptions shown below (Panel A/B), with the segments represent the expected exposure periods for each prescription (assuming each prescription lasts for 28 days).
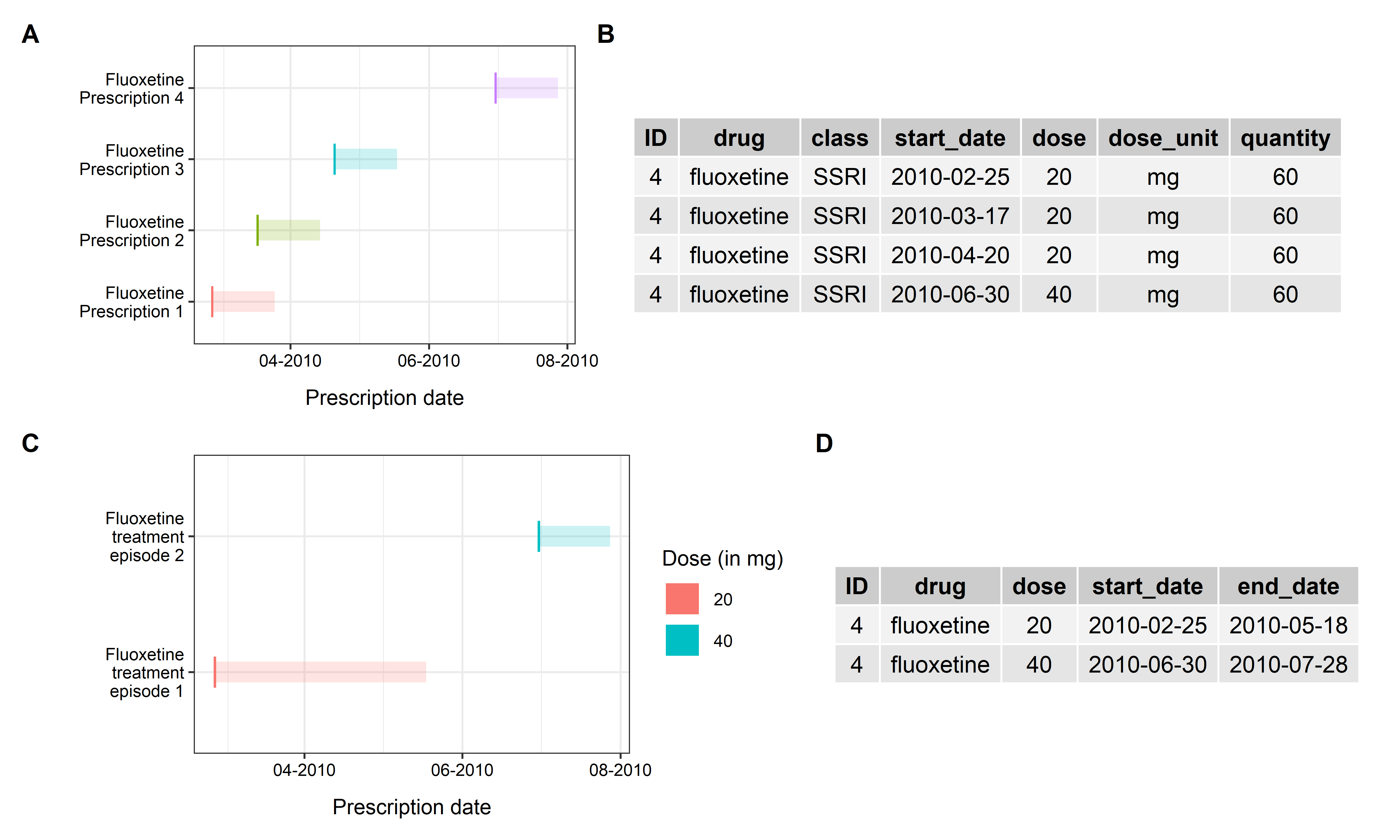
Repeated prescriptions can be defined by users based on:
- same drug name: merging all 4 prescriptions together;
- same dosage and quantity: two separate prescribing episodes will be constructed (Panel C/D).
Key Arguments to infer episode lengths
| Column | Details |
|---|---|
rx_window_days |
Gap of days between prescription dates of two consecutive prescriptions allowed to be treated as the same treatment episode, to distinguish cases of separate treatment episodes |
assume_days |
assumption on the length of final prescription (or single prescriptions) of a treatment episode, i.e.: lengths of segments in each prescriptions |
Instructions
First, build the above dataframe of 4 fluoxetine prescriptions with the following code:
Code chunk to build prescription sample
fluoxetine_rx <- data.frame(
ID = c(4, 4, 4, 4),
drug = c("fluoxetine", "fluoxetine", "fluoxetine", "fluoxetine"),
class = c("SSRI", "SSRI", "SSRI", "SSRI"),
start_date= as.Date(c("2010-02-25", "2010-03-17", "2010-04-20", "2010-06-30")),
dose = c(20, 20, 20, 40),
dose_unit = c("mg", "mg", "mg", "mg"),
quantity = c(60, 60, 60, 60),
stringsAsFactors = FALSE)Now, try to build the prescription episodes with
rx_infer(), using:
- 98 days (14 weeks) between consecutive prescriptions
(
rx_window_days) to distinguish separate treatment episodes; and - 28 days as an assumption (
assume_days) for the length of final prescriptions in the treatment episode.
Here, we are demonstrating an example of rx_infer()
below.
fluoxetine_rx_infer = rx_infer(rx_df = fluoxetine_rx, id_col = "ID",
drug_col = "drug",
date_col = "start_date",
rx_window_days = 98, assume_days = 28)Explanation on arguments
| Argument | Details |
|---|---|
rx_df |
The prescription dataframe
(fluoxetine_rx) |
id_col |
Column name that contains patient ID
(ID) |
drug_col |
Column name that contain the drug names of
prescriptions (drug) |
dose_col |
Column name that contains the drug doses of
prescriptions (NULL) |
freq_col |
Column name that contains the drug quantities /
frequencies of prescriptions (NULL) |
date_col |
Column name that contains the prescription (start)
dates of prescriptions (start_date) |
rx_window_days |
Gap of days between prescription dates of two consecutive prescriptions allowed to be treated as the same treatment episode, to distinguish cases of separate treatment episodes |
assume_days |
assumption on the length of final prescription (or single prescriptions) of a treatment episode, i.e.: lengths of segments in each prescriptions |
In rx_infer(), dose_col and
freq_col are optional arguments that specify the column
names of the respective columns, if users wish to group up the treatment
episodes by same dose and/or same dosing
frequency/quantity.
If users prefer to group up repeated prescriptions just by same drug
names only (like above), these arguments can be left as
NULL.
Output Dataframe
Running rx_infer() as above would return one single
prescription episode (since all of them are fluoxetine prescriptions and
close enough, within 98 days).
| ID | drug | start_date | end_date | n_epi |
|---|---|---|---|---|
| 4 | fluoxetine | 2010-02-25 | 2010-07-28 | 1 |
Column name descriptions
| Column | Details |
|---|---|
ID |
Numeric identifier for individual patients |
drug |
Name of the prescribed drug |
start_date |
Start date of the prescription episode |
end_date |
End date of the prescription episode |
n_epi |
Number of episode (for the group, same drug for this case) |
The start date of the episode would be 2010-02-25, the
prescription date of first prescription, whereas the end date of the
episode would be 2010-07-28, 28 days (specified by
assume_days) after the prescription date of the final
prescription (2010-06-30).
Customizing parameters
If users want to establish repeated prescriptions by same drug, same
dose and same frequency/quantity, these can be inputted to the
rx_infer() function as the dose_col and
freq_col argument.
A sample code is show below.
fluoxetine_rx_infer = rx_infer(rx_df = fluoxetine_rx, id_col = "ID",
drug_col = "drug", dose_col = "dose", freq_col = "quantity",
date_col = "start_date",
rx_window_days = 98, assume_days = 28)If you run rx_infer() by grouping up prescriptions by
dose / quantity, this would return 2 prescribing episodes as below.
| ID | drug | dose | quantity | start_date | end_date | n_epi |
|---|---|---|---|---|---|---|
| 4 | fluoxetine | 20 | 60 | 2010-02-25 | 2010-05-18 | 1 |
| 4 | fluoxetine | 40 | 60 | 2010-06-30 | 2010-07-28 | 1 |
It returns two prescribing episodes (as shown in above figure, Panel C) as:
| Episode | Duration |
|---|---|
| 20mg | 2010-02-15 to 2010-05-18 (28
days after the 3rd fluoxetine prescription,
2010-04-20) |
| 40mg | 2010-06-30 to 2010-07-28 (28
days after the final fluoxetine prescription,
2010-06-30) |
Any questions?
Please post questions as an issue on the T-Rx GitHub repo here.
The T-Rx package is currently under beta testing. Most functions should have adequate documentation on possible errors.
Please kindly reach out to Chris Lo (chris.lowh@kcl.ac.uk) for feedback on documentation.
![]()