Phenotyping Example (UK Biobank)
Introduction
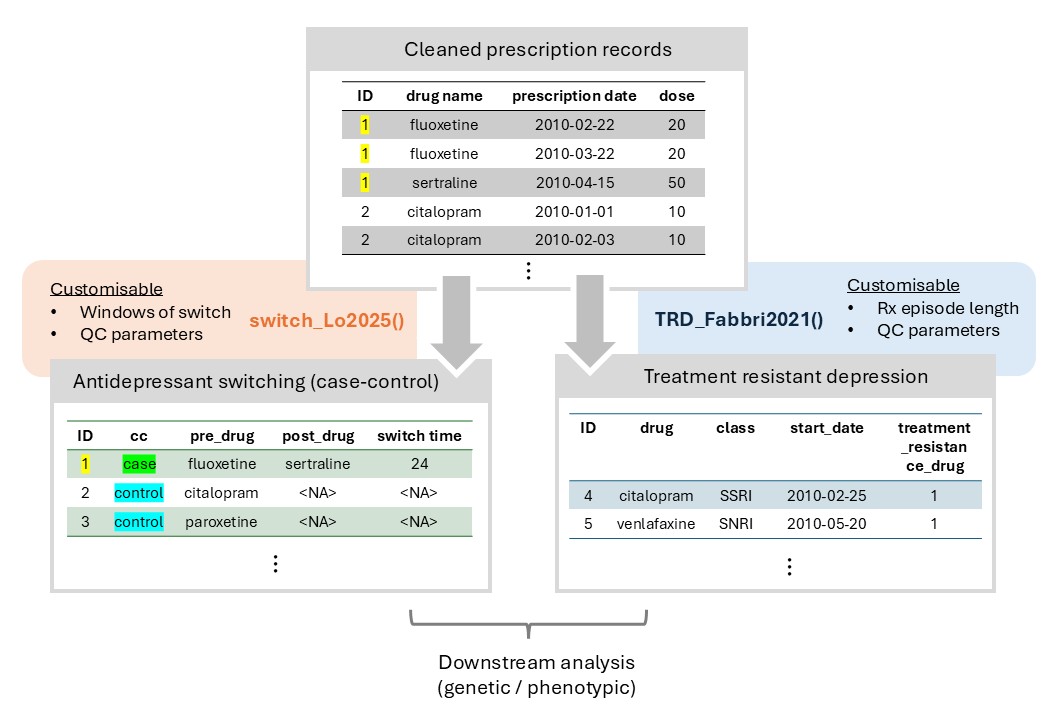
Figure 1: Overview of the phenotyping module
Overview
The Phenotyping Module of T-Rx primarily consists of two sections:
- Instructions to apply phenotyping functions to prescription data
- Contribution of phenotypes
Sample dataframe
In this phenotyping tutorial, we utilized the data object attached
with T-Rx object, rx_demo_1 for
illustration.
It is a hypothetical prescription dataframe consists of 35 rows and 8 columns (for 4 patients), with details described below.
| Column | Details |
|---|---|
ID |
Numeric identifier for individual patients |
drug |
Name of the prescribed drug |
class |
Drug class or category (e.g., SSRI) |
start_date |
Start date of the prescription |
end_date |
End date of the prescription |
dose |
Dosage of the prescribed drug |
dose_unit |
Dosage unit of the prescribed drug |
quantity |
Quantity of the drug prescribed in the prescription |
Initialization
Load T-Rx and insepct the data object in R.
# load package
library(TRX)
# inspect sample data object (antidepressant prescriptions)
dim(antidep_ukb_rx)
# [1] 35 8Inspect prescription sample
| ID | drug | class | start_date | end_date | dose | dose_unit | quantity |
|---|---|---|---|---|---|---|---|
| 1 | fluoxetine | SSRI | 2010-02-22 | 2010-03-22 | 20 | mg | 28 |
| 1 | fluoxetine | SSRI | 2010-03-22 | 2010-04-19 | 20 | mg | 50 |
| 1 | sertraline | SSRI | 2010-04-15 | 2010-05-15 | 50 | mg | 14 |
| 1 | fluoxetine | SSRI | 2010-04-15 | 2010-05-15 | 20 | mg | 14 |
| 1 | sertraline | SSRI | 2010-05-18 | 2010-06-15 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-06-10 | 2010-07-04 | 50 | mg | 30 |
Antidepressant (SSRI) switching - switch_Lo2025()
Background and Publication
In clinical guidelines for major depressive disorder, patients are recommended to switch to a different drug upon failure to respond to the initial antidepressant.
This can be captured in prescription records by studying the switching events between antidepressants in an episode for depression, as a proxy treatment-phenotype.
Papers published using the switching algorithm:
- The Effects of CYP2C19 Genotype on Proxies of SSRI Antidepressant Response in the UK Biobank: publication
- Antidepressant switching as a proxy phenotype for drug non-response: investigating clinical, demographic and genetic characteristics: publication
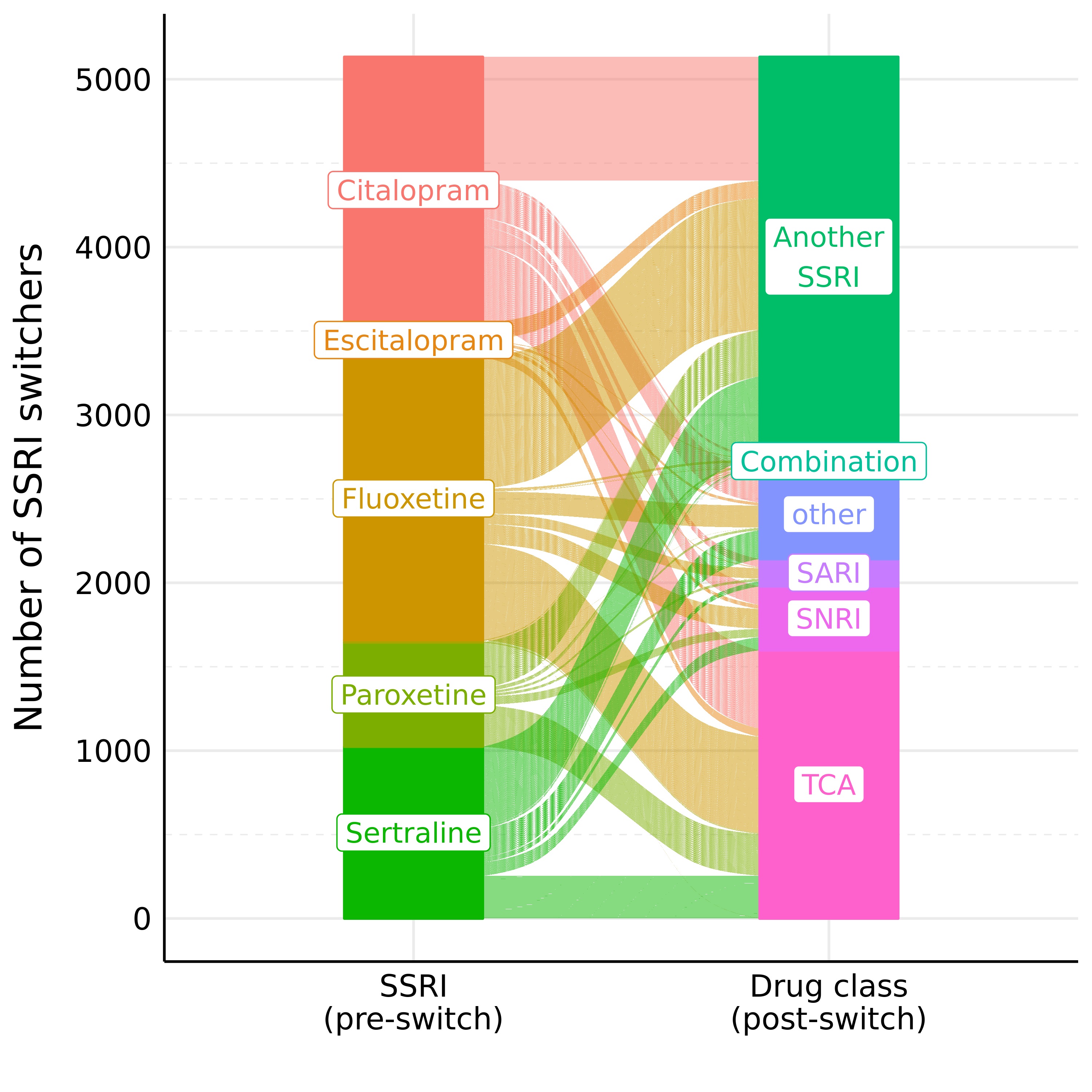
Figure 2: SSRI Switching patterns in UK Biobank (as alluvial plot)
The Switching Phenotype
The phenotyping algorithm for switching relies on gaps between prescriptions dates of two different drugs (antidepressants) with customisable options on phenotyping parameters, such as:
- Window of switch (gaps between two prescriptions of different drugs)
- Nudging window, to avoid capturing concomitant prescriptions of two drugs as switches
- Number of prescriptions of the pre-switched drug before/after the date of switch
- Total number of prescriptions of the pre-switched drug across all prescribing journeys
This approach spares us from making inferences on durations of treatment/exposure, which is often unavailable in prescription records.
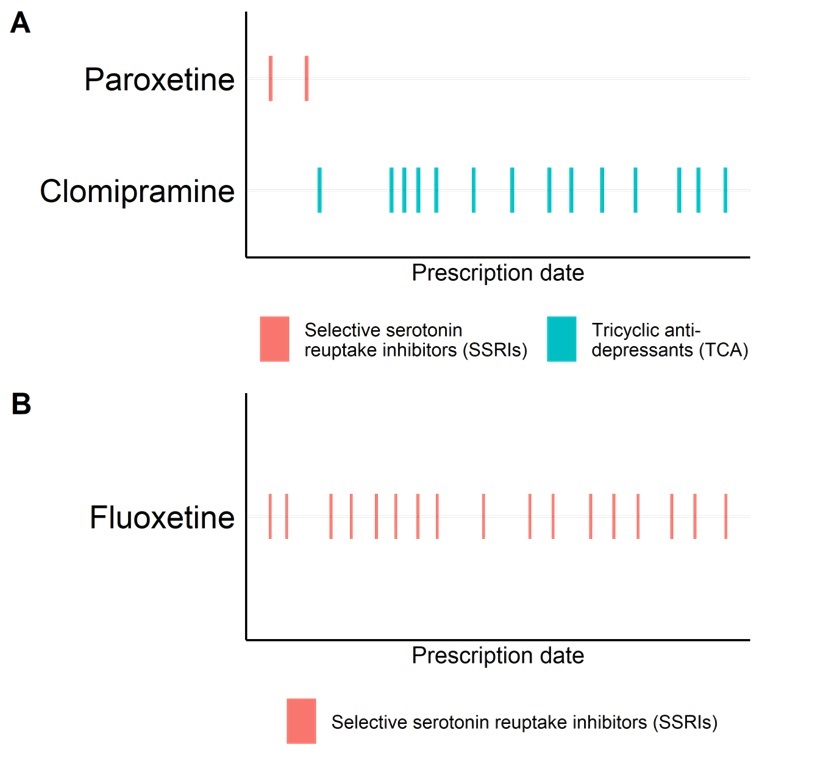
Figure 3: Example of SSRI switching
Running the function
Instructions
First, try an initial run with the switch_Lo2025()
function.
library("TRX", quietly=T)
result_switch <- switch_Lo2025(
df = rx_demo_1,
id_col = "ID", name_col = "drug", class_col = "class", date_col = "start_date",
time_switch = 90, nudge_days = 5,
pre_switch_count = 2, post_switch_count = 2, total_count = 3,
identify_controls = TRUE,
prescription_episode_days_control = 180,
consecutive_prescriptions_days_control = 30)Parameters used in the publication:
Switchers
- “switching” within a 90-day window
- 5-day of “nudging” to avoid capturing overlapping prescriptions of two different antidepressants
- pre-switch drug was prescribed:
- ≤ 3 times in all prescribing history
- ≤ 2 times before/after the switch date
Non-switchers
- ≥ 3 consecutive prescriptions of the same drug
- ≤ 30 days between consecutive prescriptions
- ≤ 180 days apart between the 3 prescriptions used to define non-switchers
Description on arguments
| Argument | Details |
|---|---|
df |
Prescription dataframe (rx_demo_1) |
id_col |
Column name for patient identifiers |
name_col |
Column name for drug names |
class_col |
Column name for drug classes |
date_col |
Column name for prescription dates |
time_switch |
Maximum allowed time (in days) between prescription dates for a switch to be considered valid |
nudge_days |
The buffer period (in days) added to remove cases of augmentation & prescription overlaps |
pre_switch_count |
Maximum number of prescriptions for a drug allowed before the switch |
post_switch_count |
Maximum number of prescriptions for a drug allowed after the switch |
total_count |
Maximum total number of prescriptions for a drug across all prescribing journeys |
identify_controls |
Whether controls need to be identified |
prescription_episode_days_control |
The maximum number of days to consider prescriptions as part of the same prescription episode |
consecutive_prescriptions_days_control |
he maximum number of days between consecutive prescriptions to qualify as consistent prescriptions |
class_filter |
Character vector specifying drug classes to filter switchers and non-switchers |
What the function does in the background
switch_Lo2025() is divided into three
sections (please refer to the source code if useful):
wrangle_switch_Lo2025(): Concatenates prescriptions from long format to wide format.switch_qc_Lo2025(): From the concatenated data frame, identify quality-controlled switching events.case_control_assemble_Lo2025(): Identify switchers (cases) and non-switchers (controls) and assemble into a phenotype dataframe, with switching details.
Running switch_Lo2025() should return a dataframe with 3
rows (3 patients):
| pt_id | cc | pre_drug | pre_class | switch_time | post_drug | post_class | index_date |
|---|---|---|---|---|---|---|---|
| 1 | case | fluoxetine | SSRI | 52 | sertraline | SSRI | 2010-02-22 |
| 4 | case | citalopram | SSRI | 63 | fluoxetine | SSRI | 2009-12-24 |
| 3 | control | paroxetine;paroxetine;paroxetine;paroxetine;paroxetine | SSRI;SSRI;SSRI;SSRI;SSRI | NA | NA | NA | 2010-01-01 |
Column descriptions
| Column | Details |
|---|---|
pt_id |
Unique patient identifier |
cc |
Case-control status (case for switchers, control for non-switchers) |
pre_drug |
Name of the drug the patient switched from |
pre_class |
Class of the drug the patient swtiched from |
switch_time |
Time to switch (in days) |
post_drug |
Name of the drug the patient switched to |
post_class |
Class of the drug the patient swtiched to |
index_date |
Index date (first prescription date of pre-switch drug) |
Switchers
We can refer back to the switchers (Example: pt_id = 1)
to see if the functions are capturing patterns as expected.
pt_1 = rx_demo_1[rx_demo_1$ID == "1",]
print(pt_1)Looking into the dataframe you would see patient 1
(pt_id = 1) received two prescriptions of fluoxetine on
2010-02-22 and 2010-03-22.
Afterwards, sertraline is prescribed on 2010-04-15, 52
days after the first prescription of fluoxetine
(2010-02-22).
These are shown in the switch_result dataframe with the
details of switching:
- pre_drug: fluoxetine
- post_drug: sertraline
- switch_time: 52
- index_date: 2010-02-22 (date of first fluoxetine prescription)
| ID | drug | class | start_date | end_date | dose | dose_unit | quantity |
|---|---|---|---|---|---|---|---|
| 1 | fluoxetine | SSRI | 2010-02-22 | 2010-03-22 | 20 | mg | 28 |
| 1 | fluoxetine | SSRI | 2010-03-22 | 2010-04-19 | 20 | mg | 50 |
| 1 | sertraline | SSRI | 2010-04-15 | 2010-05-15 | 50 | mg | 14 |
| 1 | fluoxetine | SSRI | 2010-04-15 | 2010-05-15 | 20 | mg | 14 |
| 1 | sertraline | SSRI | 2010-05-18 | 2010-06-15 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-06-10 | 2010-07-04 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-07-02 | 2010-08-01 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-07-29 | 2010-08-26 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-08-24 | 2010-09-23 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-09-27 | 2010-10-25 | 50 | mg | 30 |
| 1 | sertraline | SSRI | 2010-10-25 | 2011-01-23 | 50 | mg | 90 |
| 1 | sertraline | SSRI | 2011-01-20 | 2011-02-17 | 50 | mg | 30 |
Non-switchers
We can also have a look at a sample for non-switchers
(pt_id = 3), where the patient receives 5 consecutive
prescriptions of paroxetine, starting from 2010-01-01.
You might also notice the strings were concatenated for non-switchers
/ controls (separated by ;). This is to keep convenience if
users wants to trace how many prescriptions the non-switcher received in
the prescription episode.
| ID | drug | class | start_date | end_date | dose | dose_unit | quantity | |
|---|---|---|---|---|---|---|---|---|
| 15 | 3 | paroxetine | SSRI | 2010-01-01 | 2010-01-29 | 20 | mg | 28 |
| 16 | 3 | paroxetine | SSRI | 2010-01-26 | 2010-02-25 | 20 | mg | 28 |
| 17 | 3 | paroxetine | SSRI | 2010-02-28 | 2010-03-30 | 20 | mg | 28 |
| 18 | 3 | paroxetine | SSRI | 2010-03-26 | 2010-04-16 | 20 | mg | 28 |
| 19 | 3 | paroxetine | SSRI | 2010-04-17 | 2010-05-17 | 20 | mg | 28 |
To convert to a cleaned format, run the following:
switch_class = switch_class %>%
rowwise() %>%
mutate(pre_drug = unique(unlist(strsplit(pre_drug, split=";"))),
pre_class = unique(unlist(strsplit(pre_class, split=";")))) %>%
ungroup()This should return a dataframe with the pre-switch drugs and classes of non-switchers being cleaned into one drug.
Tweaking parameters
Switching window
If users wanted to capture patients who switched in a shorter window,
change the time_switch parameter.
result_switch <- switch_Lo2025(
df = rx_demo_1,
id_col = "ID", name_col = "drug", class_col = "class", date_col = "start_date",
time_switch = 50, nudge_days = 5,
pre_switch_count = 2, post_switch_count = 2, total_count = 3,
identify_controls = TRUE,
prescription_episode_days_control = 180,
consecutive_prescriptions_days_control = 30)Resultant dataframe
| pt_id | cc | pre_drug | pre_class | switch_time | post_drug | post_class | index_date |
|---|---|---|---|---|---|---|---|
| 1 | case | fluoxetine | SSRI | 52 | sertraline | SSRI | 2010-02-22 |
| 3 | control | paroxetine;paroxetine;paroxetine;paroxetine;paroxetine | SSRI;SSRI;SSRI;SSRI;SSRI | NA | NA | NA | 2010-01-01 |
| 4 | control | citalopram;citalopram;citalopram;fluoxetine;fluoxetine;fluoxetine;venlafaxine;venlafaxine;venlafaxine;venlafaxine;amitriptyline;perphenazine | SSRI;SSRI;SSRI;SSRI;SSRI;SSRI;SNRI;SNRI;SNRI;SNRI;TCA;atypical_antipsychotic | NA | NA | NA | 2009-12-24 |
You would notice the following changes:
Patient 1 is no longer classified as a case, as the switch from fluoxetine to sertraline occurred 52 days after the initial fluoxetine prescription.
Instead, patient 1 becomes controls for fluoxetine / sertraline, because he/she does not switch and received ≥ 3 consecutive prescriptions of both drugs.
Like above, we reserve the flexibility for users to define the drug used as “control” episode, which can be fluoxetine / sertraline with different index dates (first date of prescription).
Below summarises the codes on how to convert these controls:
First control episode
result_switch = result_switch %>%
rowwise() %>%
mutate(pre_drug = unique(unlist(strsplit(pre_drug, split=";")))[1],
pre_class = unique(unlist(strsplit(pre_class, split=";")))[1]) %>%
ungroup()Control episode for a specific drug (sertraline for example), under development
Dataframe without controls
Users might have alternative algorithms for identifying non-switchers (controls), and particularly be interested in switchers only.
switch_Lo2025() offers an argument of
not identifying non-switchers, so the resultant dataframe will only
contain switcher information.
This can be done by setting identify_controls argument
to FALSE.
Example code
switchers_only <- switch_Lo2025(
df = rx_demo_1,
id_col = "ID", name_col = "drug", class_col = "class", date_col = "start_date",
time_switch = 90, nudge_days = 5,
pre_switch_count = 2, post_switch_count = 2, total_count = 3,
identify_controls = FALSE)Resultant dataframe
| pt_id | cc | pre_drug | pre_class | switch_time | post_drug | post_class | index_date |
|---|---|---|---|---|---|---|---|
| 1 | case | fluoxetine | SSRI | 52 | sertraline | SSRI | 2010-02-22 |
| 4 | case | citalopram | SSRI | 63 | fluoxetine | SSRI | 2009-12-24 |
Treatment-resistant depression - TRD_Fabbri2021()
Background and Publication
Treatment-resistant depression (TRD) is usually defined as lack of response to at least two antidepressants.
In prescription records, this can be captured by identifying individuals with MDD having at least two switches between different antidepressant drugs (independently from the class).
Original papers published to define TRD:
Genetic and clinical characteristics of treatment-resistant depression using primary care records in two UK cohorts: publication
Transcriptome-wide association study of treatment-resistant depression and depression subtypes for drug repurposing: publication
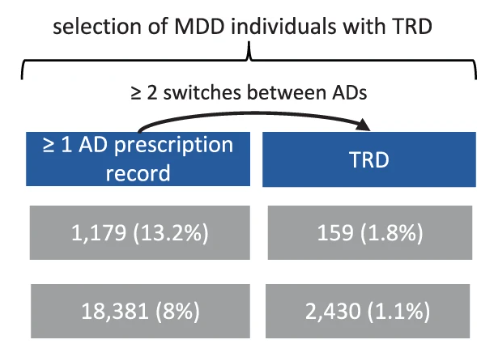
Figure 4: Descriptive figure for TRD (Fabbri et al., 2021)
The TRD Phenotype
According the original authors, TRD is defined as individuals with:
- ≥ 2 switches in antidepressants;
- ≥ 6 weeks for each drug prescribed;
- ≤ 14 weeks between prescriptions of two consecutive drugs (switching from one drug to another);
- ≤ 14 weeks between two consecutive prescriptions (regardless of whether same drug or not).
Note: In the original paper, the authors also consider two prescriptions of the same antidepressant as the same treatment episodes if the prescription dates were ≤ 26 weeks apart.
Running the function
Instructions
First, try an initial run with the TRD_Fabbri2021()
function.
library("TRX", quietly=T)
result_TRD <- TRD_Fabbri2021(
df = rx_demo_1,
id_col = "ID", name_col = "drug", class_col = "class", date_col = "start_date",
week_prescription_episode = 26,
week_poor_compliance = 14,
week_adequate_treatment = 6,
week_non_suspend = 14)Note: The default parameters were those used in Fabbri et al., 2022.
Description on arguments
| Argument | Details |
|---|---|
df |
Prescription dataframe (rx_demo_1) |
id_col |
Column name for patient identifiers |
name_col |
Column name for drug names |
class_col |
Column name for drug classes |
date_col |
Column name for prescription dates |
week_prescription_episode |
An integer specifying the maximum gap (in weeks) between prescriptions of the same antidepressant to be considered the same prescribing episode |
week_poor_compliance |
An integer specifying the gap (in weeks) between prescriptions for identifying cases of poor compliance |
week_adequate_treatment |
An integer specifying the minimum duration (in weeks) for a prescription episode to be considered adequate |
week_non_suspend |
An integer specifying the maximum gap (in weeks) between prescription of two different drugs in a sequence to still be considered in TRD |
TRD individuals
Running TRD_Fabbri2021() should yield a dataframe of two
rows of the same individual (ID = 4) as below.
TRD phenotype sample
| ID | drug | class | start_date | between_drugs_weeks | prescription_episode_drug | adequate_prescr_period | treatment_resistance_drug |
|---|---|---|---|---|---|---|---|
| 4 | fluoxetine | SSRI | 2010-04-20 | 4.571429 | 7.714286 | 1 | 1 |
| 4 | venlafaxine | SNRI | 2010-08-28 | 4.714286 | 14.000000 | 1 | 1 |
Column descriptions
| Column | Details |
|---|---|
id_col |
Unique patient identifiers |
name_col |
Drugs that each patient switched before reaching TRD (as N rows) |
class_col |
Drug classes for drugs that each patient switched before reaching TRD (as N rows) |
date_col |
The date of the final prescription of that drug before it is switched |
between_drugs_weeks |
Intervals (in weeks) between the final prescription of the drug and the previous prescription of the same drug |
prescription_episode_drug |
Duration (in weeks) of the prescribing episode for the drug |
adequate_prescr_period |
Logical indicator of whether the prescription met the
criteria for adequate duration (according to
week_adequate_treatment argument) |
treatment_resistance_drug |
Logical indicator (phenotype) of whether the patient was identified as having treatment-resistant depression |
Contributing your phenotypes
We are very keen to expand our library of phenotyping algorithm for reproducibility and scalability.
Please kindly reach out to Chris Lo (chris.lowh@kcl.ac.uk) or learn more here.
Troubleshooting
Please post questions as an issue on the T-Rx GitHub repo here.
The T-Rx package is currently under beta testing. Most functions should have adequate documentation on possible errors.
Please kindly reach out to Chris Lo (chris.lowh@kcl.ac.uk) for feedback on documentation.
![]()