T-Rx - homepage
![]()
A Toolbox for reproducible processing of
prescription (Rx)
records
Based at King’s College London, T-Rx is a R package that aims to improve the accessibility of complex electronic health records, by developing reproducible algorithms to extract, impute prescription details and creating phenotypes.
![]()
Overview
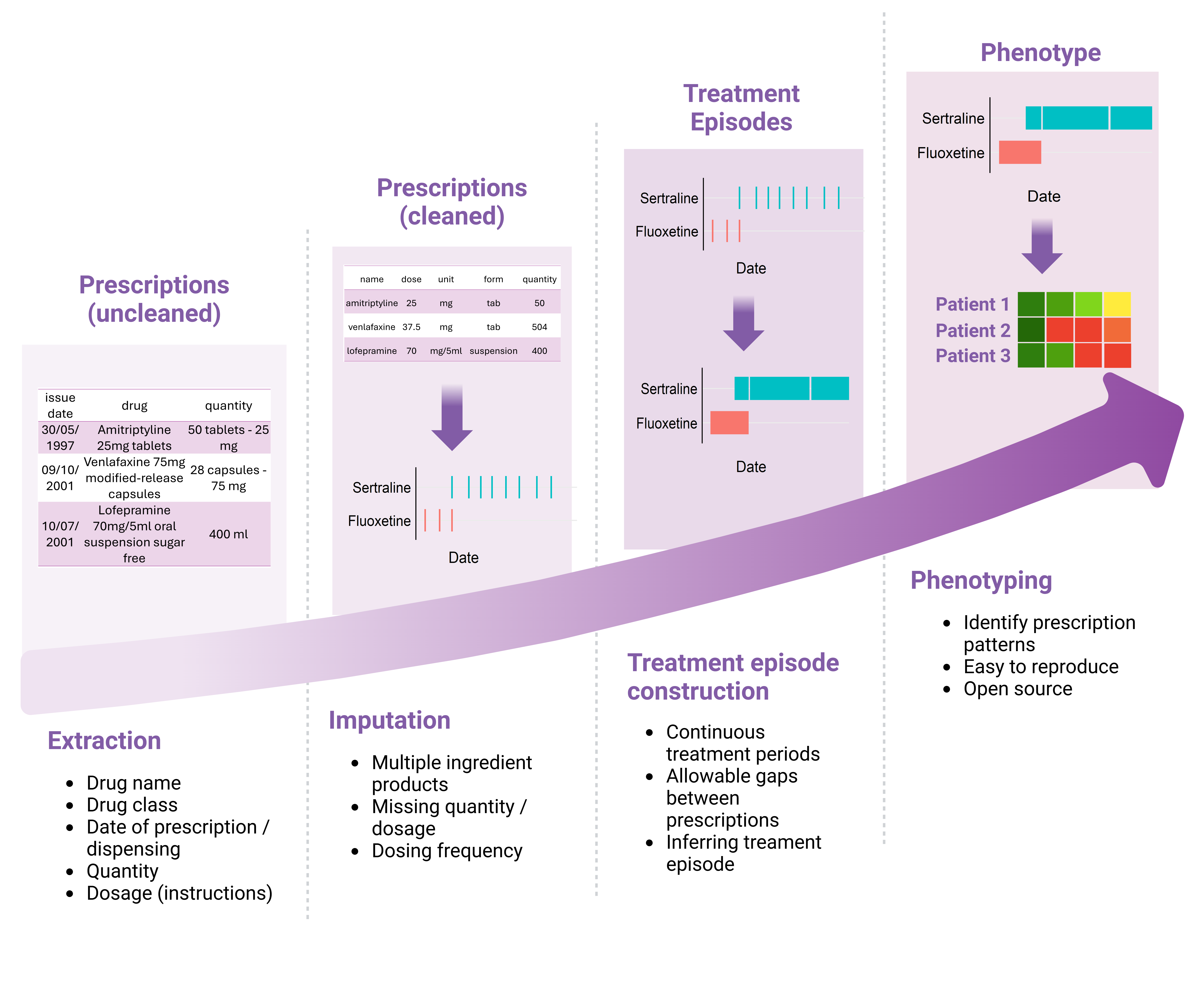
T-RX provides solutions for conversion from raw prescription records to:
- extract & impute prescription details using regular expression patterns
- host coding rules for treatment-related proxy phenotypes as reproducible and open-sourced functions;
- provide a toolbox of functions for wrangling EHR data, such as inferring treatment episodes.
Extraction and Imputation Module
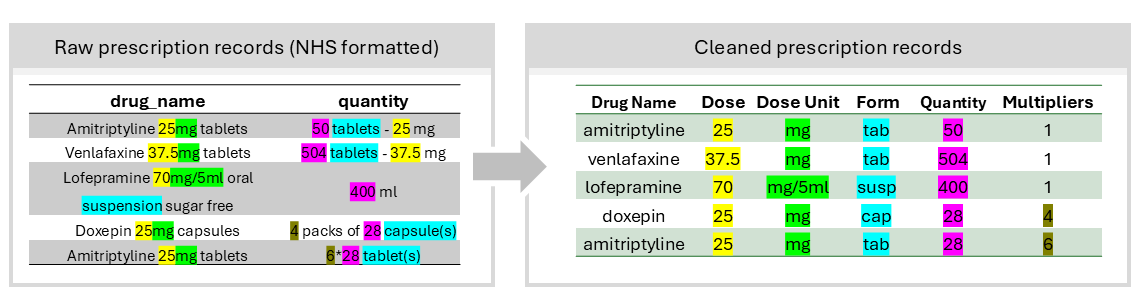
Figure 1: Overview of prescription extraction by regular expression patterns, with UK Biobank as example
Exposure Ascertainment Module
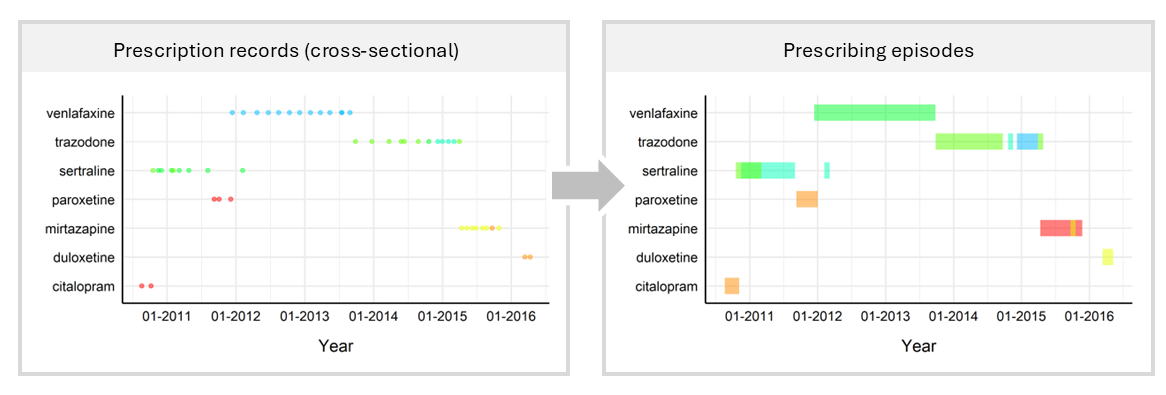
Figure 2: Overview of converting prescriptions to longitudinal periods of exposure
Phenotyping Module
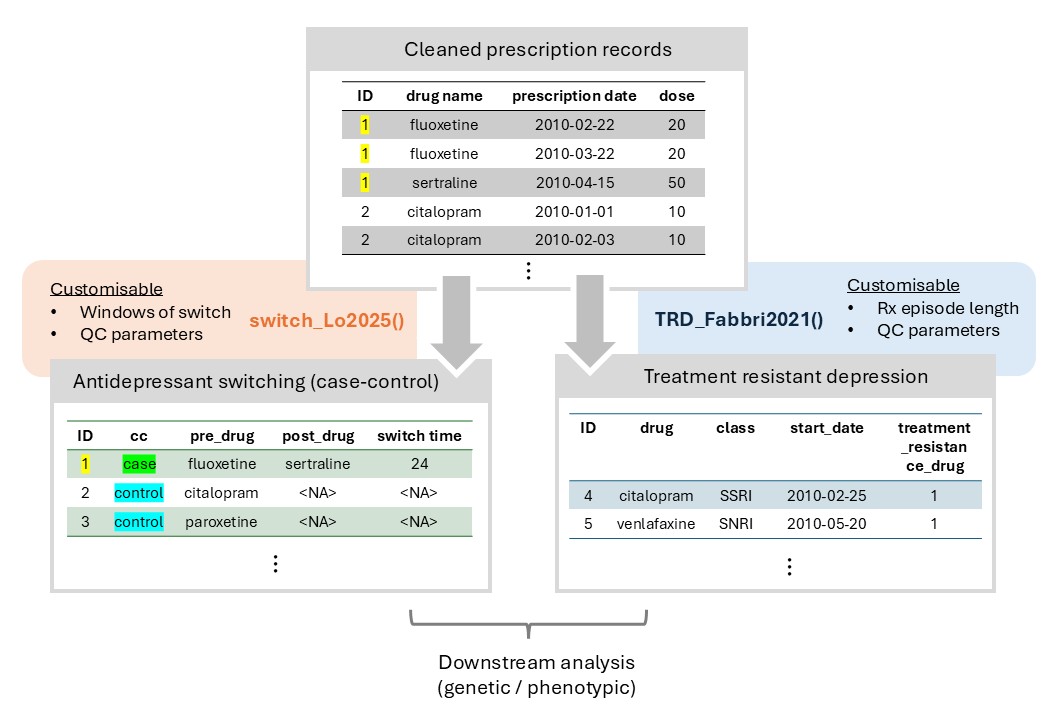
Figure 3: Overview of phenotyping module, using antidepressant switching and treatment-resistant depression as examples
AMBER Study
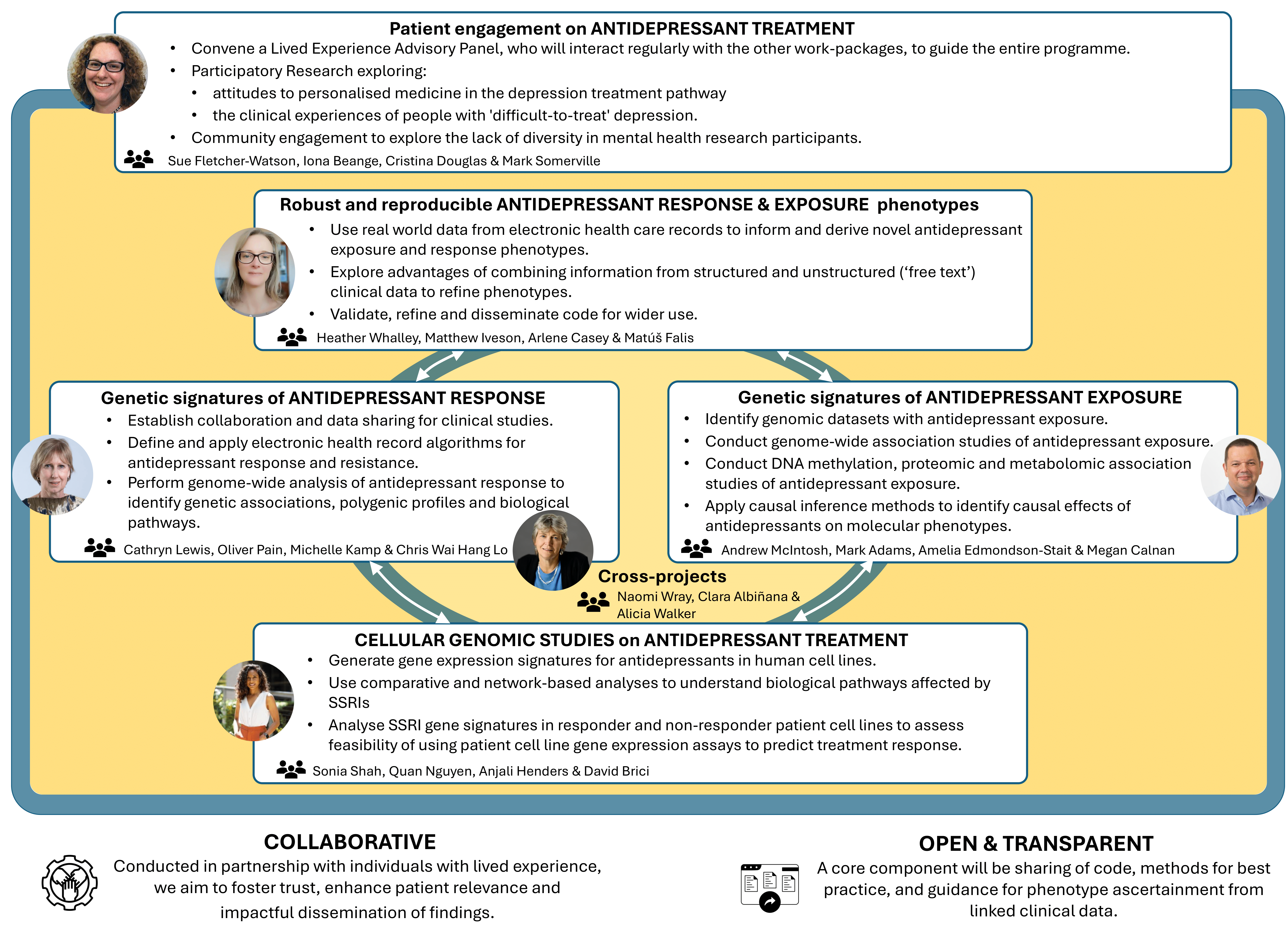
The AMBER study (Antidepressant Medications: Biology, Exposure & Response) is a Wellcome-funded Mental Health Award that aims to identify the causal determinants of antidepressant response (2023-2028).
The AMBER team integrates clinical, genomic, cellular and patient-participatory research to provide insights into antidepressant action and response, enhance understanding of drug mechanisms and biological pathways, and develop predictive models building towards personalised prescribing to improve patient outcomes.
![]()